The Role of Donor-Derived Cell-Free DNA in the Identification of Injury in Kidney Allografts with Antibody-Mediated Rejection or De Novo DSA: A Pilot Study
Huanxi Zhang1, Longshan Liu1, Chunting Zheng3, Xirui Li1, Qian Fu1, Jun Li1, Qun Su1, Huiting Huang1, Mingzhi Ye3, Changxi Wang1,2.
1Organ transplant center, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou, P.R. China; 2Guangdong Provincial Key Laboratory on Organ Donation and Transplant Immunology, Guangzhou, P.R. China; 3BGI-Guangzhou Medical Laboratory, BGI-Shenzhen, Guangzhou, P.R. China
Objectives: To evaluate the capacity of donor-derived cell-free DNA (dd-cfDNA) in the identification of injury in kidney allografts with antibody-mediated rejection or de novo DSA without histological lesions.
Methods: Blood specimens were collected in kidney allograft recipients with either antibody-mediated rejection (AMR) or de novo DSA (dnDSA) without histological lesions. The recipients with stable serum creatinine and negative DSA were enrolled into the control group. The plasma level of dd-cfDNA in recipients was accurately measured using a non-donor-derived cfDNA transplant dynamics approach, which is implemented by genotyping with only genomic DNA from a recipient’s blood cells by targeted capture next-generation sequencing assay in a mini-screen SNP array and calculating donor fraction of cfDNA from the plasma of the recipient by extra-low depth whole genome sequencing.
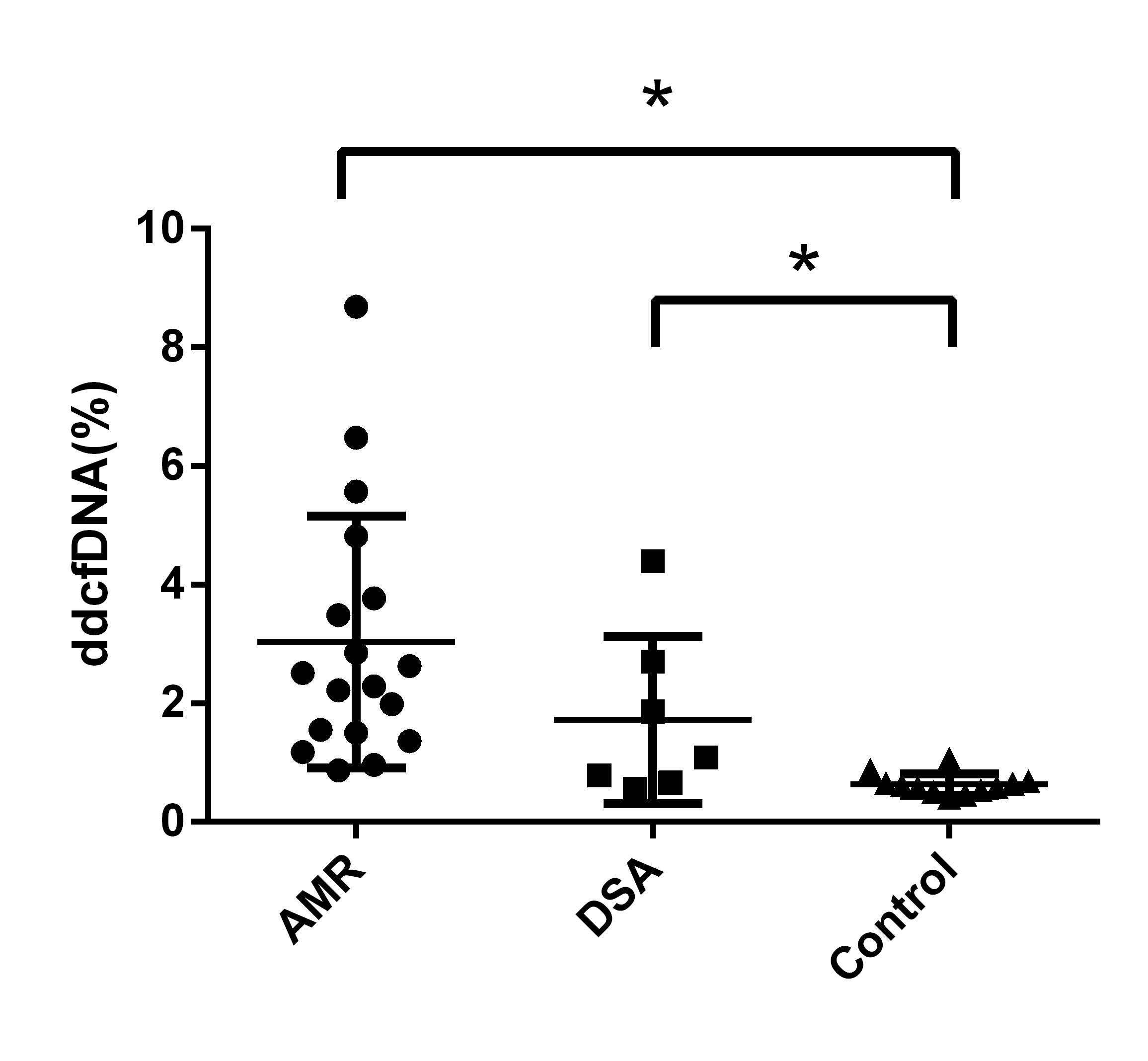
Results: Thirty-seven recipients were enrolled for analysis (18 AMR, 7 dnDSA and 12 controls). A significantly increased level of dd-cfDNA was identified in AMR group or dnDSA group compared with control group [figure1] (AMR 3.04%±2.12% vs Control 0.63%±0.18%, P<0.05; dnDSA 1.72%±1.41% vs Control 0.63%±0.18%, P<0.05; ANOVA P<0.01). The receiver operating characteristic area under the curve (AUC) was 0.97 (95% confidence interval [95%CI], 0.96 to 1.00) and 0.84 (95%CI, 0.65 to 1.00) respectively for the discrimination of AMR or dnDSA from the control group. Positive/negative predictive values and positive/negative likelihood ratios for AMR at a cutoff of 1.0% dd-cfDNA were 94.1%/84.6% and 10.7/0.12, respectively. There were 4 samples (4/7, 57%) with dd-cfDNA level >1% in dnDSA group, which may reflect injury to the allografts.
Conclusions and Discussion: Donor-derived cfDNA may be used to assess allograft injury in AMR or recipients with dnDSA but no histological lesions. Donor-derived cfDNA levels (cutoff = 1%) can discriminate AMR from stable recipients. Roy D. Bloom et al. also reported the capacity of dd-cfDNA for discriminating AMR from no allograft injury or some other injuries (T cell-mediated rejection, interstitial fibrosis/tubular atrophy and acute tubular necrosis, etc.) 1. Based on this result, we identified 4 dnDSA(+) recipients with dd-cfDNA level >1%. We will closely monitor these recipients to investigate whether histological lesions will occur sooner compared with those with dd-cfDNA level <1%. The study is still ongoing and particularly more recipients with dnDSA but no histological lesions will be enrolled to identify the correlation between dd-cfDNA level and the prognosis of these recipients.
Reference:
1. Bloom RD, Bromberg JS, Poggio ED, et al. Cell-Free DNA and Active Rejection in Kidney Allografts. J Am Soc Nephrol. 2017;28(7):2221-2232