Analysis of Differentially Expressed Proteins in Hepatocellular Carcinoma
Angeles Baquerizo1, Margaret Simonian2, Catherine Frenette1, Randolph Schaffer1, Mary Nelson1, Jonathan Fisher1, Julian Whiteledge2, Bahar Madani1, Paul Pockros1, Christopher Marsch1.
1Scripps Center for Cell and Organ Transplantation, Scripps Health, La Jolla, CA, United States; 2Mass Spectrometry, UCLA, Los Angeles, CA, United States
Introduction: Hepatocellular carcinoma (HCC), the third leading cause of cancer-related mortality worldwide, has limited treatment options; therefore, the importance of developing new and innovative therapeutic strategies. The field of proteomics has emerged as a powerful tool to identify changes in protein expression in HCC patients.
Objectives: Identify differentially expressed proteins in tissue biopsies of HCC patients who underwent liver transplantation.
Material & Methods: The protein expression in paraffin-embedded liver biopsies of 24 HCC liver transplant patients (12 Poor Differentiated/Vascular Invasion -Poor-, 12 Well/Moderate Differentiated -Well-) was assessed by Mass Spectrometry (LC/MS) . Five non-tumor liver biopsies were used as Controls (Ctr). The expression ratios of abundances were calculated (1 ± 0.25) and p-values were determined using two-tailed Student's t-test. Relevant demographics and clinical characteristics of the patients were also collected (age, sex, total tumor volume, number of tumors, peak AFP pre-surgery, vascular invasion, MELD, tumor differentiation, Child-Pugh, tumor distribution).
Results: The preliminary results are shown in Table 1. The explorative, quantitative proteome analyses identified over 4,000 proteins in HCC biopsies. Although we didn't achieve statistically significance differences given the small number of patients, we identify 56 proteins overexpressed (>1.5 fold) in poor differentiated HCC compare to Controls. These proteins were differentially expressed in well differentiated HCC vs poor differentiated HCC.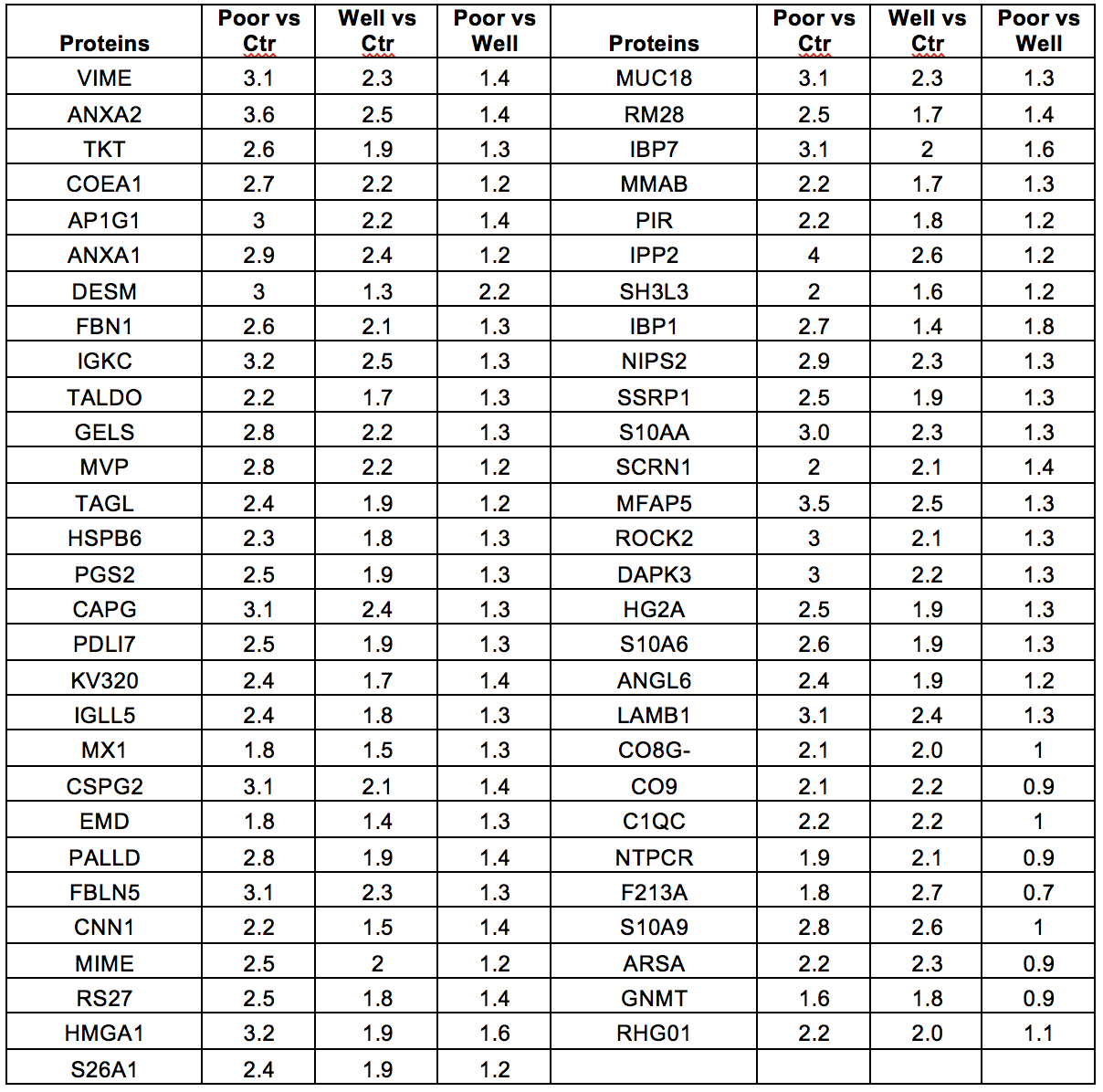
Conclusions: We identified proteins differentially expressed in poor differentiated HCC. These results may have implications as prognostic biomarkers and future targets for immunotherapy.
