Transcriptional Factor FOXP3 in Renal Allograft Biopsies Predicts Tolerance in Non-Human Primates
Masatoshi Matsunami1, Tetsu Oura1, Benjamin A. Adam2, Michael Mengel2, Ivy A. Rosales1, Rex-Neal Smith1, David Schoenfeld1, A. Benedict Cosimi1, Robert B. Colvin1, Tatsuo Kawai1.
1Department of Surgery, Center for Transplantation Sciences, Massachusetts General Hospital, Harvard Medical School, Boston, MA, United States; 2Department of Laboratory Medicine and Pathology, University of Alberta, Edmonton, AB, Canada
NIH NHP Transplantation Tolerance Cooperative Study Group.
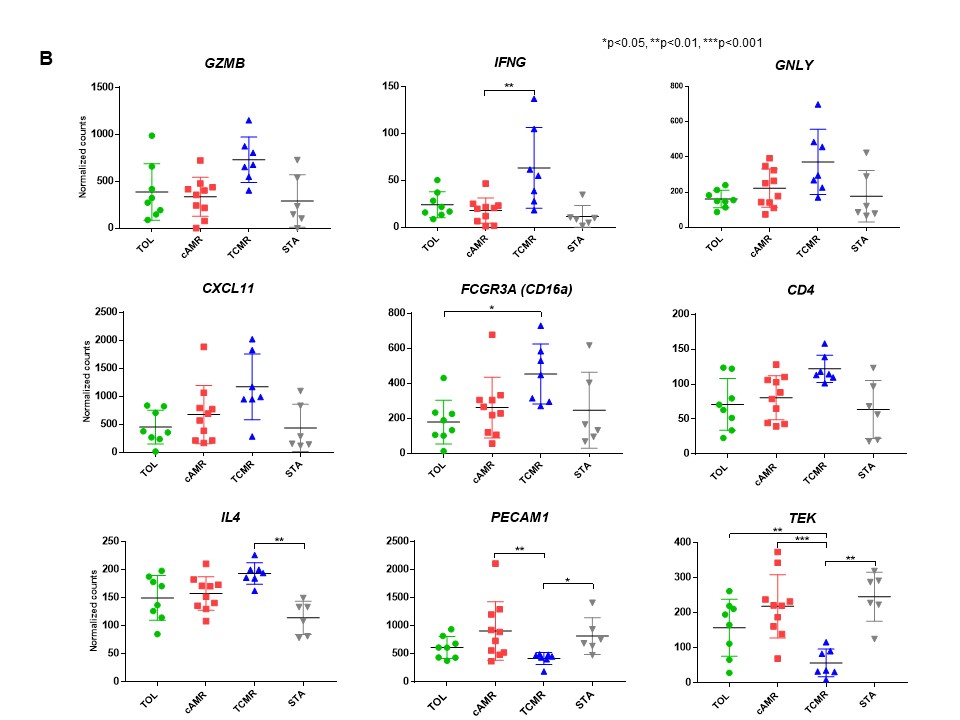
Identification of reliable biomarkers for tolerance is critically important in transplantation. We reasoned that intragraft transcripts may be more revealing than those in peripheral blood leukocytes if tolerance is mediated in part by events in the graft. Using the NanoString nCounter platform, we retrospectively studied 256 kidney allograft formalin-fixed paraffin-embedded (FFPE) serial samples taken from non-human primate recipients after combined kidney and bone marrow transplantation (CKBMT) to measure 49 genes, which were previously reported significantly associated with tolerance or rejection. We found dynamic time-dependent kinetics of intragraft biomarkers, which indicated the timing of analysis is critical to interpret the results (Figure 1).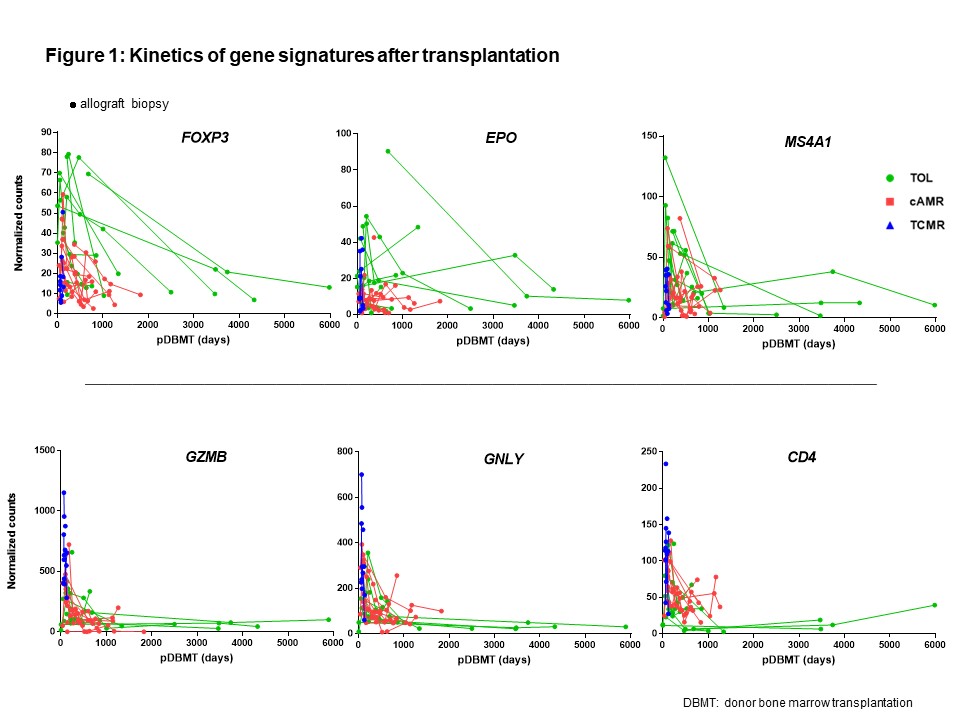 To identify biomarkers that can predict the transplant outcome at the early post-transplant period, we focused on analyzing mRNAs in biopsies taken between 30 and 120 days after CKBMT when no rejection was yet observed. The recipients that subsequently achieved tolerance (TOL, n=11, allograft survival>1995 ± 557 days) showed significantly higher mRNA expression of FOXP3, BCL2, GATA3 and RPS6KB1 compared with biopsies from recipients that later developed chronic antibody-mediated rejection (cAMR, n=11, 850 ± 134 days) or T cell mediated rejection (TCMR, n=9, 109 ± 22 days). In contrast, transcripts of inflammatory cytokines or adaptive immunity (IFNG, CXCL11, FCGR3A, GNLY, GZMB, IL4 and CD4) were higher in TCMR, while endothelium associated transcripts, such as PECAM1 and TEK, were higher in cAMR (Figure 2A and 2B).
To identify biomarkers that can predict the transplant outcome at the early post-transplant period, we focused on analyzing mRNAs in biopsies taken between 30 and 120 days after CKBMT when no rejection was yet observed. The recipients that subsequently achieved tolerance (TOL, n=11, allograft survival>1995 ± 557 days) showed significantly higher mRNA expression of FOXP3, BCL2, GATA3 and RPS6KB1 compared with biopsies from recipients that later developed chronic antibody-mediated rejection (cAMR, n=11, 850 ± 134 days) or T cell mediated rejection (TCMR, n=9, 109 ± 22 days). In contrast, transcripts of inflammatory cytokines or adaptive immunity (IFNG, CXCL11, FCGR3A, GNLY, GZMB, IL4 and CD4) were higher in TCMR, while endothelium associated transcripts, such as PECAM1 and TEK, were higher in cAMR (Figure 2A and 2B).
 The ROC curve analyses revealed that intragraft FOXP3 mRNA alone reliably differentiate TOL from both cAMR and TCMR, and that ratios of FOXP3 to several inflammatory cytokine signatures showed better AUC to differentiate TOL from TCMR (Figure 3).
The ROC curve analyses revealed that intragraft FOXP3 mRNA alone reliably differentiate TOL from both cAMR and TCMR, and that ratios of FOXP3 to several inflammatory cytokine signatures showed better AUC to differentiate TOL from TCMR (Figure 3).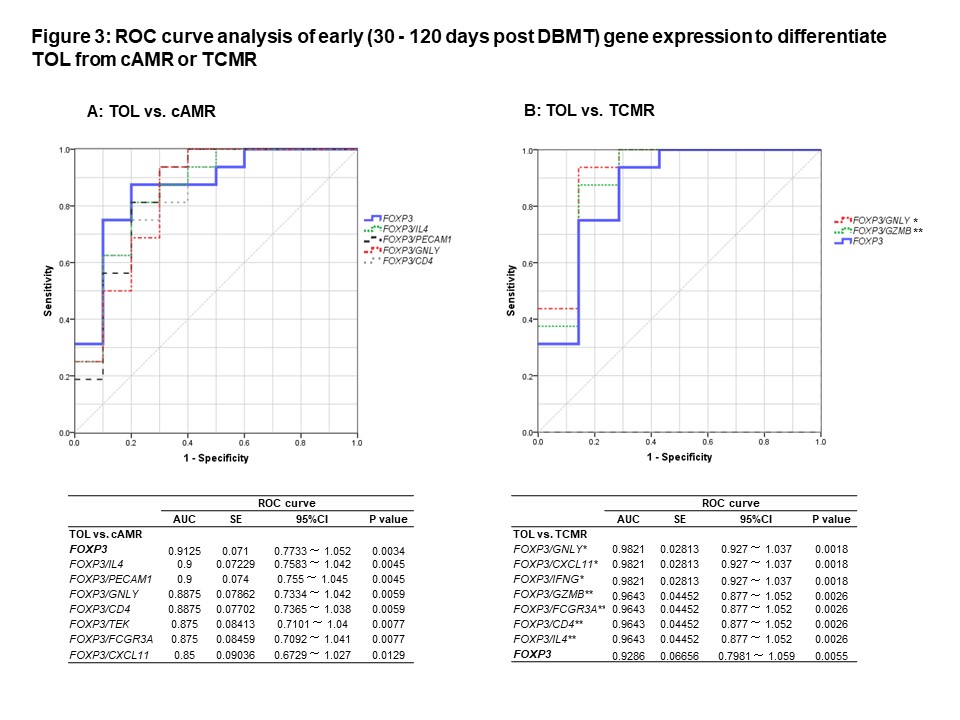 The significance of intragraft FOXP3 for tolerance was validated by the prospective study with 11 CKBMT recipients. We conclude that intragraft mRNA transcripts, especially FOXP3, measured early post transplantation can serve as biomarkers to reliably predict subsequent tolerance induction via the mixed chimerism approach.
The significance of intragraft FOXP3 for tolerance was validated by the prospective study with 11 CKBMT recipients. We conclude that intragraft mRNA transcripts, especially FOXP3, measured early post transplantation can serve as biomarkers to reliably predict subsequent tolerance induction via the mixed chimerism approach.
Grant 5U19AI102405.
