
Plasma Donor-Derived Cell-Free DNA Quantification by massively multiplex PCR Distinguishes Kidney Transplant Acute Rejection
Tara Sigdel1, Felipe Archila2, Samantha Navarro2, Bernhard Zimmermann2, Solomon Moshkevich2, Minnie Sarwal1.
1Dept of Surgery, University of California San Francisco, San Francisco, CA, United States; 2Natera, Inc., San Carlos, CA, United States
Introduction: Plasma donor-derived cell-free DNA (dd-cfDNA) has been implicated as a noninvasive marker for transplant (tx) rejection. dd-cfDNA evaluation requires differentiation of donor/recipient DNA by sequencing; recent studies have amplified hundreds of target SNPs to detect active rejection in kidney allografts with 59.3% sensitivity and 84.7% specificity. We measure thousands of informative SNPs to assess dd-cfDNA with high accuracy in a selected cohort of kidney tx patients having contemporaneous tx biopsies scored for presence and type of Banff-graded T cell-/antibody-mediated–rejection (TCMR/ABMR) and borderline rejection (BL).
Materials and Methods: 292 unique plasma samples from 187 unique patients were categorized as stable (STA; n=73), acute rejection (AR; n=52), other injury (OI; n=85), or BL (n=82), and processed by massively multiplex PCR targeting 13,392 SNPs. Cross-sectional samples were obtained from AR and OI patients (other causes of graft dysfunction were drug toxicity [n=18], acute tubular necrosis [n=2], BK nephritis [n=4], chronic allograft nephropathy [CAN; n=57], and tx glomerulpathy [n=3]). AR was scored by Banff for TCMR (n=95), ABMR (n=37), and BL. 41 patients contributed 3–4 samples each (146 samples total) over 12–24 months for longitudinal assessment. dd-cfDNA performance was evaluated by ROC with inclusion of eGFR in the prediction model. Calculations were determined using 95% confidence.
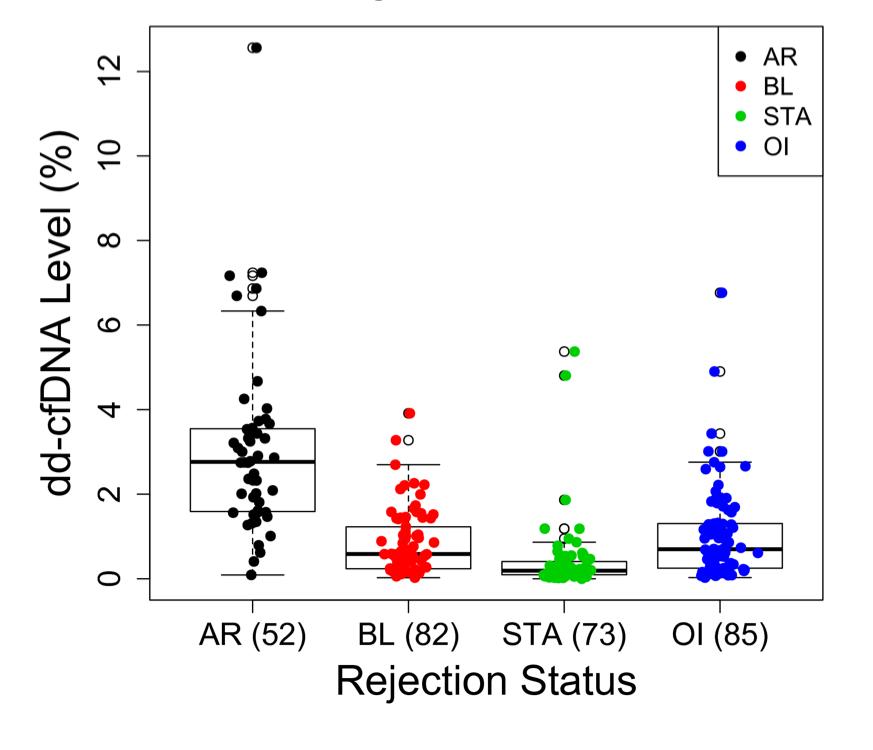
Results: dd-cfDNA circulatory burden was significantly higher in AR (3.075±2.136%) compared to STA (0.428±0.851%; p<0.0001) and OI (1.051±1.112%; p<0.0001) (Figure 1). dd-cfDNA was also higher in Banff-confirmed AR over BL rejection (0.834 ±0.765%; p<0.0001), with no difference in burden observed for TCMR and ABMR (3.003 ±2.292% and 3.185 ±1.931%, respectively; p=0.5203).
To compare dd-cfDNA to eGFR score, samples with available eGFR score were used (STA, n=7; AR, n=52). Using a cutoff of >1% dd-cfDNA, AR was detected with 91.8% sensitivity (CI 80.4–97.7) and 100% specificity (CI 59–100). Area under the curve (AUC) of 0.985 showed strong AR detection power of dd-cfDNA. Using a logistic regression integrating both dd-cfDNA and eGFR with a >50% probability cutoff, classification of samples was 100% accurate. Estimated CI for sensitivity and specificity were (92.7–100) and (59–100), respectively, with AUC of 1 (compared with AUC of 0.79 using eGFR alone).
Discussion: The novel SNP-based mmPCR assay enabled rapid detection of dd-cfDNA without need for sequencing or laborious analytics. Irrespective of rejection type, the assay observed a threshold for STA patients and an exponential increase in kidney injury burden in CAN and BL rejection with much greater burden in AR; taken together, these data suggest that combined dd-cfDNA and eGFR markers can accurately assess AR risk in kidney tx recipients.
Conclusion: This technology may provide a less invasive and more sensitive approach to monitoring the health of kidney allografts.
Figure 1. Relationship of Plasma dd-cfDNA Levels and Graft Rejection Status
Natera, Inc..
