
Molecular Assessment of Heart Transplant Biopsies: Emergence of the Injury Dimension
Michael Parkes1, Jeff Reeve1, Daniel Kim1, Peter Macdonald3, Arezu Aliabadi4, Johannes Goekler4, Andreas Zuckermann4, Patrick Bruneval6, Alexandre Loupy6, Luciano Potena2, Marisa Crespo-Leiro5, Martin Cadeiras7, Eugene Depasquale7, Mario Deng7, Jon Kobashigawa8, Philip Halloran1.
1University of Alberta, Edmonton, AB, Canada; 2Università di Bologna, Bologna, Italy; 3Victor Chang Cardiac Research Institute, Darlinghurst, Australia; 4Medical University of Vienna, Vienna, Austria; 5Universitario de A Coruña, A Coruña, Spain; 6Hôpital Necker, Paris, France; 7University of California Los Angeles, Los Angeles, CA, United States; 8Cedars-Sinai Medical Center, Beverley Hills, CA, United States
INTERHEART (ClinicalTrails.gov NCT02670408).
Introduction: We previously developed a molecular diagnostic system for assessment of rejection phenotypes in endomyocardial biopsies (EMB) based on expression of rejection-associated transcripts (RAT) derived in kidney (JHLT 36:1192, 2017). This system used archetypal analysis to identify 3 rejection-related molecular phenotypes among the biopsies (A1NoRejection, A2TCMR, A3ABMR), with each biopsy assigned scores relating them to the molecular phenotypes. We now explored whether there was another dimension beyond rejection that reflected acute parenchymal injury.
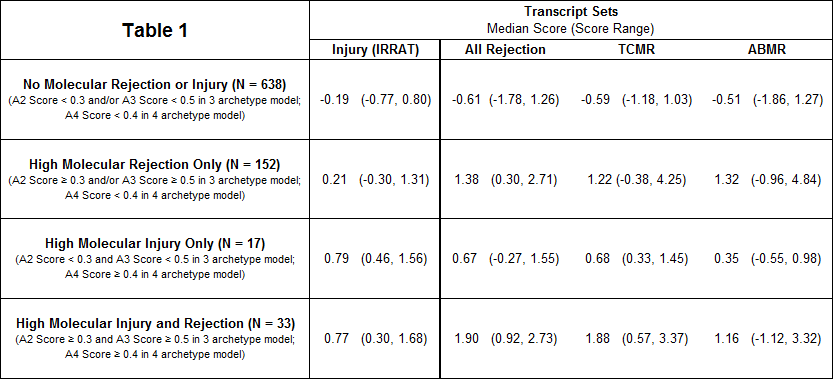
Materials & Methods: 889 single-piece EMBs from 462 heart transplant recipients at 8 centres in North America, Europe, and Australia were analyzed on Affymetrix microarrays. We used 2 methods to assess injury: archetypal analysis to assign groups, and expression of previously defined injury-repair transcripts (IRRAT). EMBs with A2TCMR scores ≥ 0.3 and A3ABMR scores ≥ 0.5 in the previously published 3 archetype model were designated “molecular rejection.” EMBs with A4 scores ≥ 0.4 in a new 4 archetype model were designated “molecular injury.” The rejection and injury designations were not mutually exclusive.
Results: EMBs characterized by archetypal analysis using 4 rather than 3 archetypes were distributed by principal component analysis based on RAT expression (Figure 1). PC1 reflected rejection, and PC2 reflected ABMR vs. TCMR (Figure 1A). The new group (A4) had an “acute injury” phenotype that was most apparent in PC3 (Figure 1B).
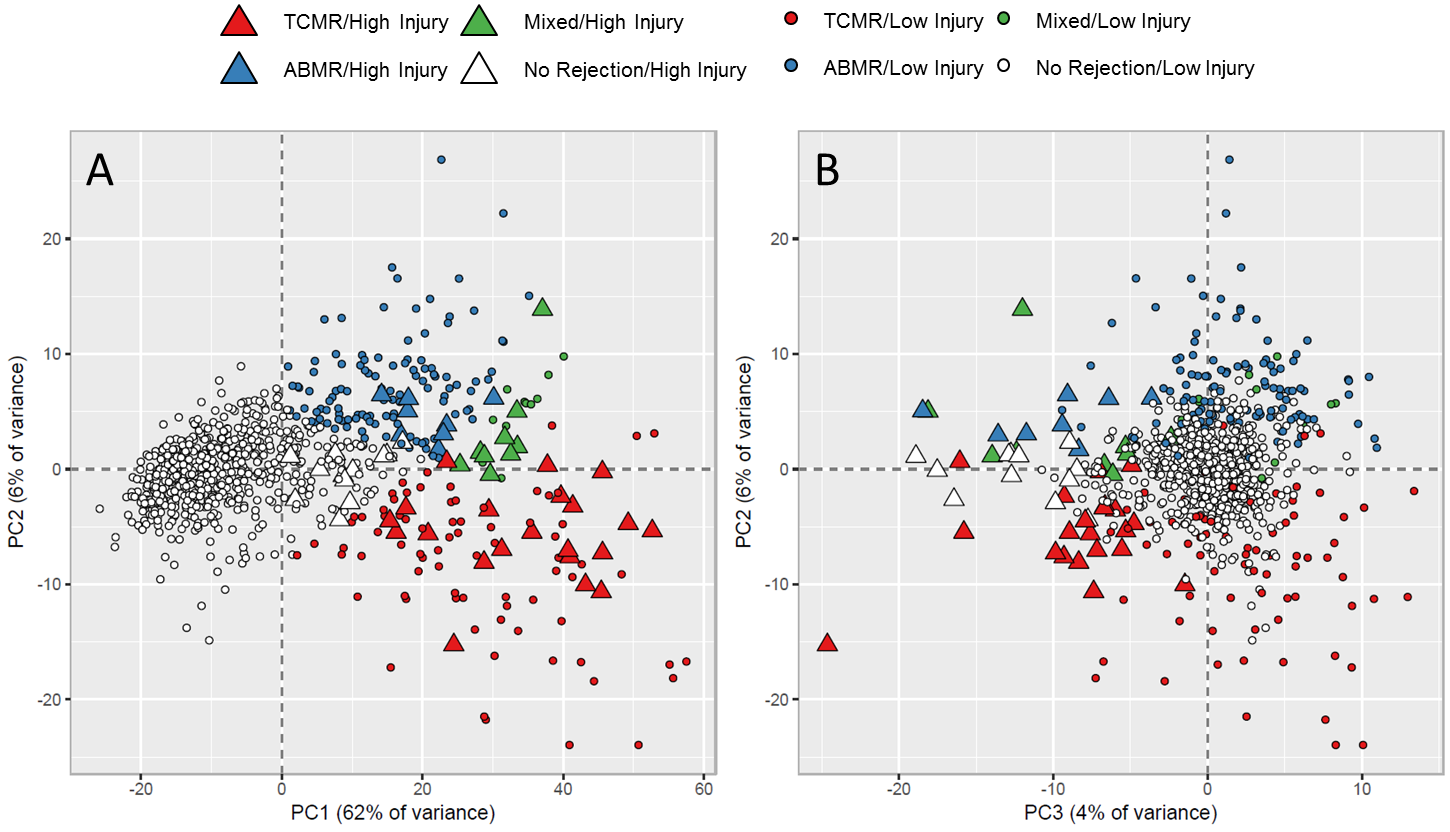
A4 had high expression of macrophage transcripts and IRRAT. The median IRRAT score in EMBs with high injury and low rejection scores was high compared to relatively normal biopsies (0.79 vs. -0.19) (Table 1). Median IRRAT scores in EMBs with high rejection and low injury were also elevated, albeit to a lesser extent (0.21 vs. -0.19). The expression of rejection transcripts was somewhat elevated in acute injury without rejection, reflecting overlap between inflammatory processes activated in rejection and non- rejection-related injury.
Many A4 EMBs were taken very early post-transplant (median time 30 days), probably reflecting injury induced in donation/implantation. Histology sometimes misdiagnosed rejection in A4 EMBs because of the macrophage infiltration: of 17 EMBs with high molecular injury and no molecular rejection, 9 were called rejection by histology.
Conclusion: Unsupervised analysis with 4 archetypes discovered a new group, acute injury, with high expression of macrophage and injury transcripts and early time post-transplant. Thus hearts often have an early acute injury phenotype that is inflamed, more so than in kidney transplants. Some of these biopsies have molecular rejection but some do not. Some with no molecular rejection are called rejection by histology, apparently reflecting injury-induced inflammation. We conclude that a molecular approach that independently assesses rejection and injury is needed for EMBs.
Transcriptome Sciences, Inc..
